Downloadable files
The ScPCA Portal download packages include gene expression data, a QC report, and associated metadata for each processed sample.
Gene expression data is available as either SingleCellExperiment objects (.rds files) or AnnData objects (.hdf5 files).
These files are delivered as a zip file.
When you uncompress the zip file, the root directory name of your download will include the date you accessed the data on the ScPCA Portal.
We recommend you record this date in case there are future updates to the Portal that change the underlying data or if you need to cite the data in the future (see How to Cite for more information).
Please see our CHANGELOG for a summary of changes that impact downloads from the Portal.
For all downloads, sample folders (indicated by the SCPCS prefix) contain the files for all libraries (SCPCL prefix) derived from that biological sample.
Most samples only have one library that has been sequenced.
For multiplexed sample libraries, the sample folder name will be an underscore-separated list of all samples found in the library files that the folder contains.
Note that multiplexed sample libraries are only available as SingleCellExperiment objects, and are not currently available as AnnData objects.
See the FAQ section about samples and libraries for more information.
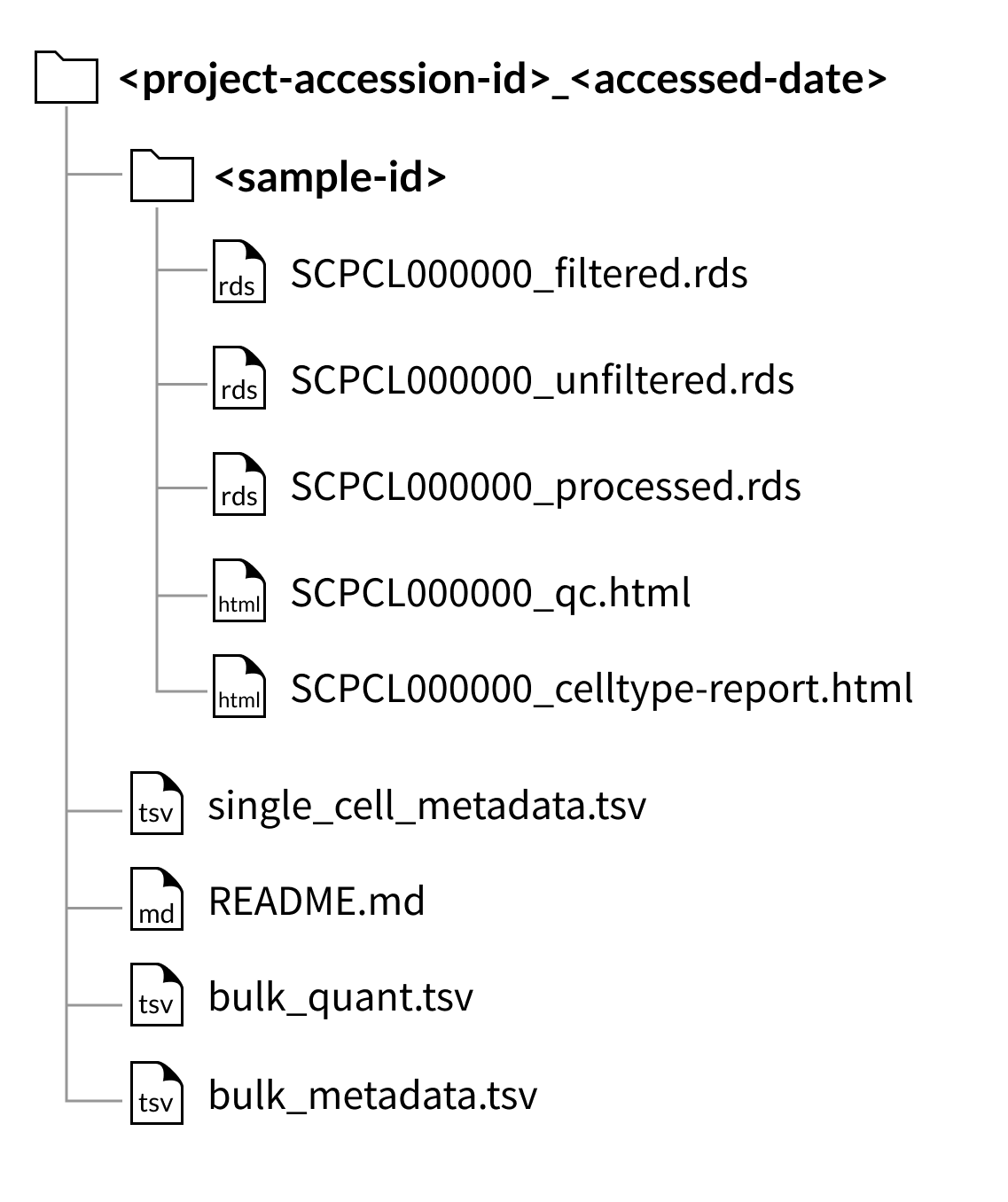
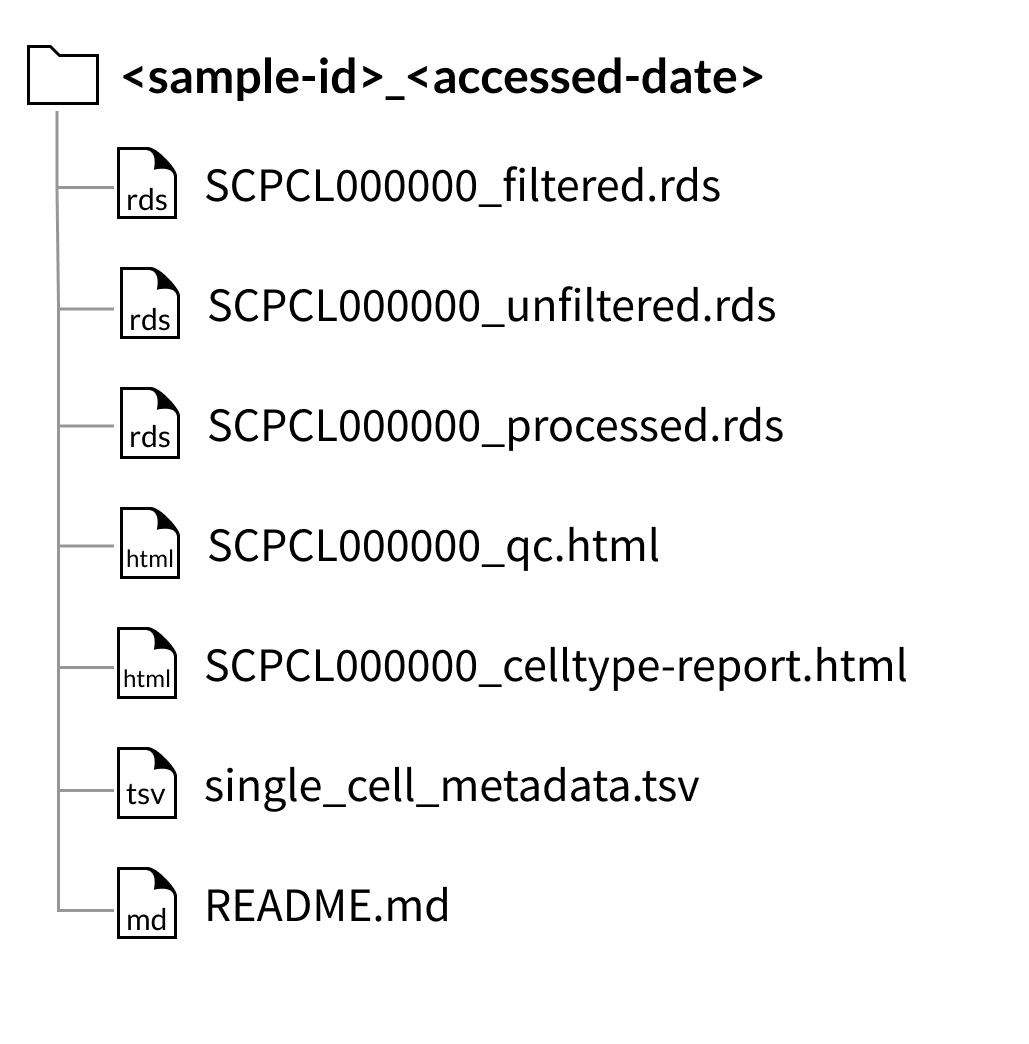
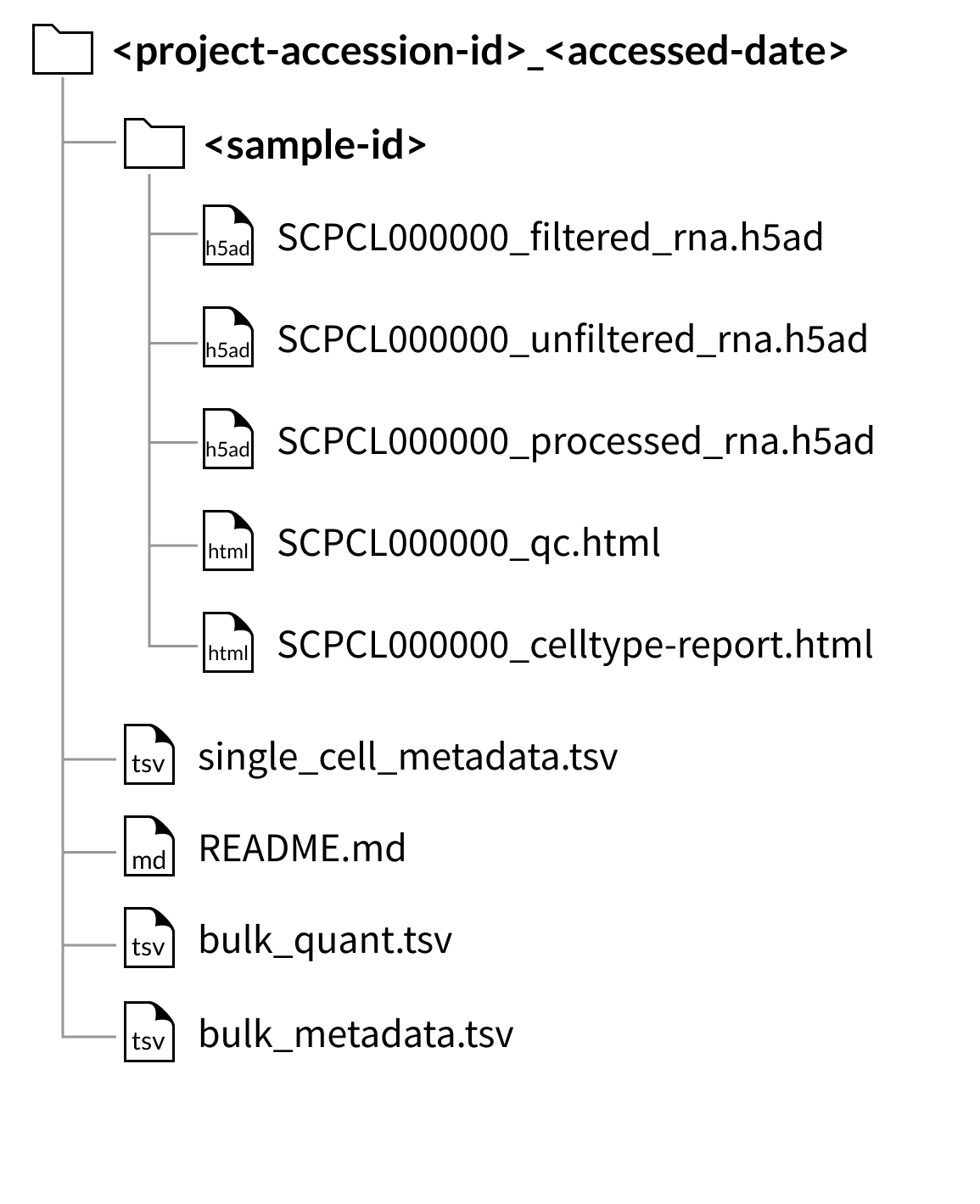
The files shown below will be included with each library (example shown for a library with ID SCPCL000000):
An unfiltered counts file:
SCPCL000000_unfiltered.rdsorSCPCL00000_unfiltered_rna.hdf5,A filtered counts file:
SCPCL000000_filtered.rdsorSCPCL00000_filtered_rna.hdf5,A processed counts file:
SCPCL000000_processed.rdsorSCPCL00000_processed_rna.hdf5,A quality control report:
SCPCL000000_qc.html,A supplemental cell type report:
SCPCL000000_celltype-report.html
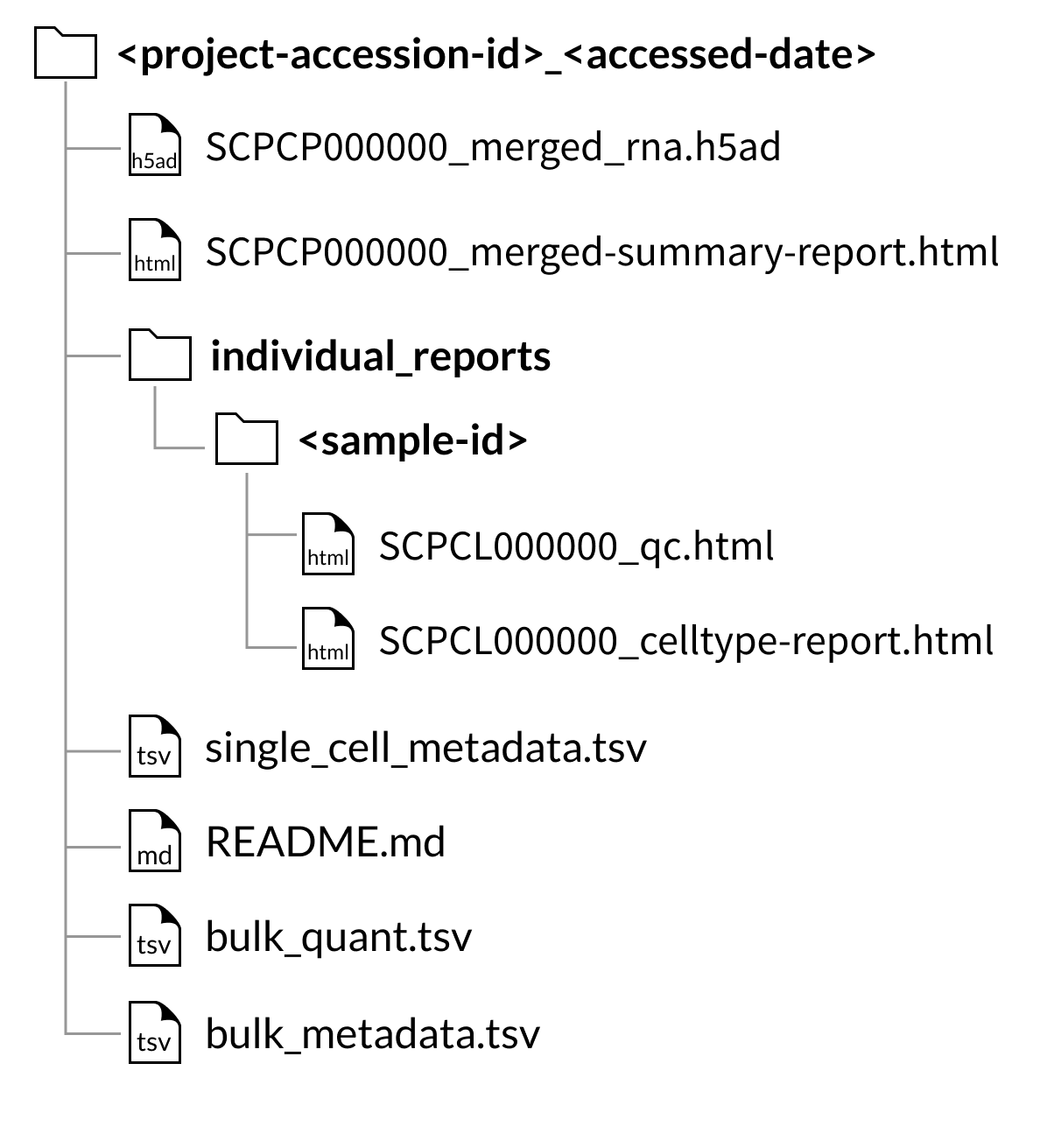
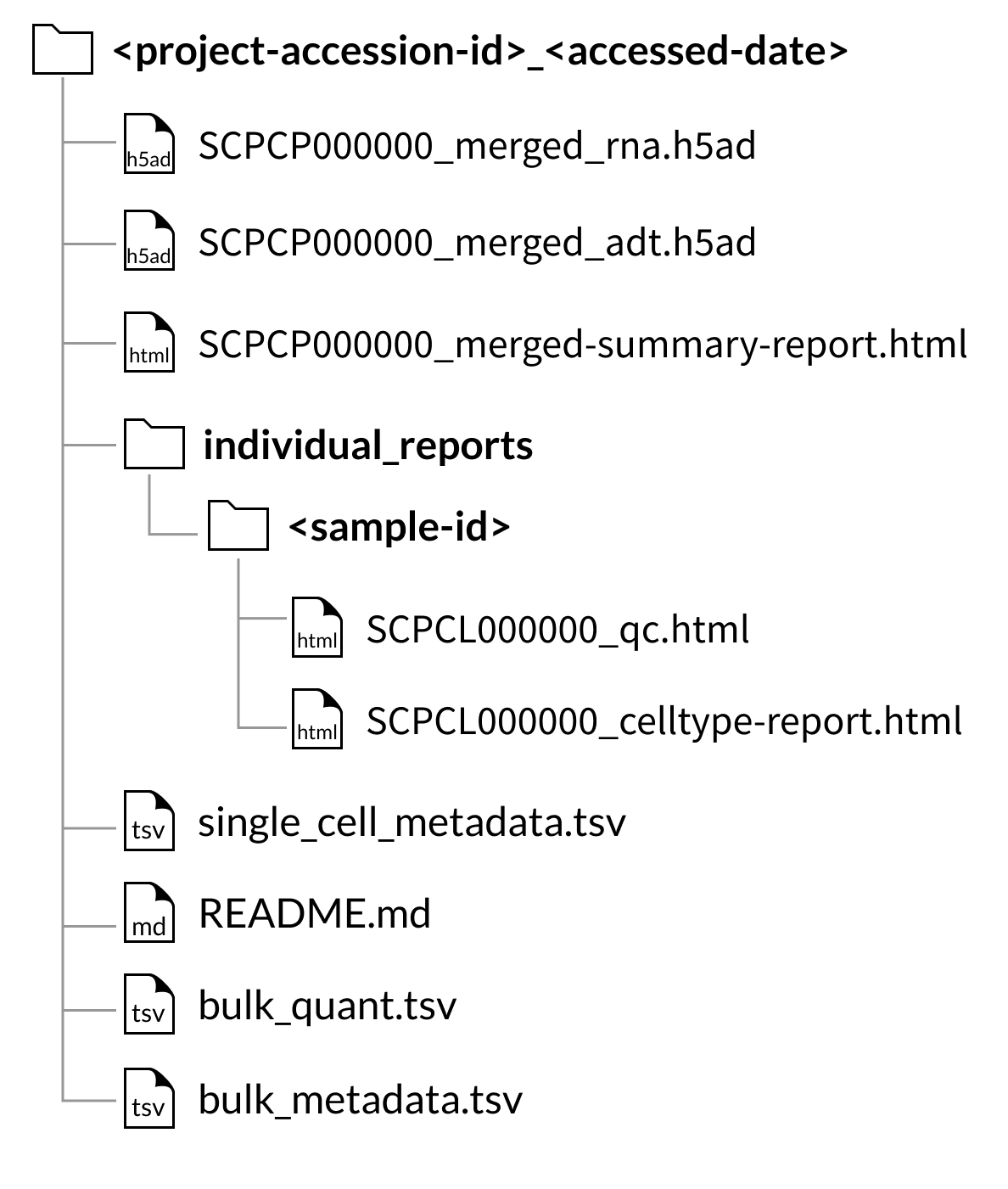
Every download also includes a single single_cell_metadata.tsv file containing metadata for all libraries included in the download.
If downloading a project containing bulk RNA-seq data, two tab-separated value files, bulk_quant.tsv and bulk_metadata.tsv, will be included in the project download.
The bulk_quant.tsv file contains a gene by sample matrix (each row a gene, each column a sample) containing raw gene expression counts quantified by Salmon.
The bulk_metadata.tsv file contains associated metadata for all samples with bulk RNA-seq data.
See also processing bulk RNA samples.
The folder structure within the zip file is determined by whether individual samples or all samples associated with a project are selected for download. Note that if a sample selected for download contains a spatial transcriptomics library, the files included will be different than pictured below. See the description of the Spatial transcriptomics output section below.
SingleCellExperiment downloads
Download folder structure for project downloads:
Download folder structure for individual sample downloads:
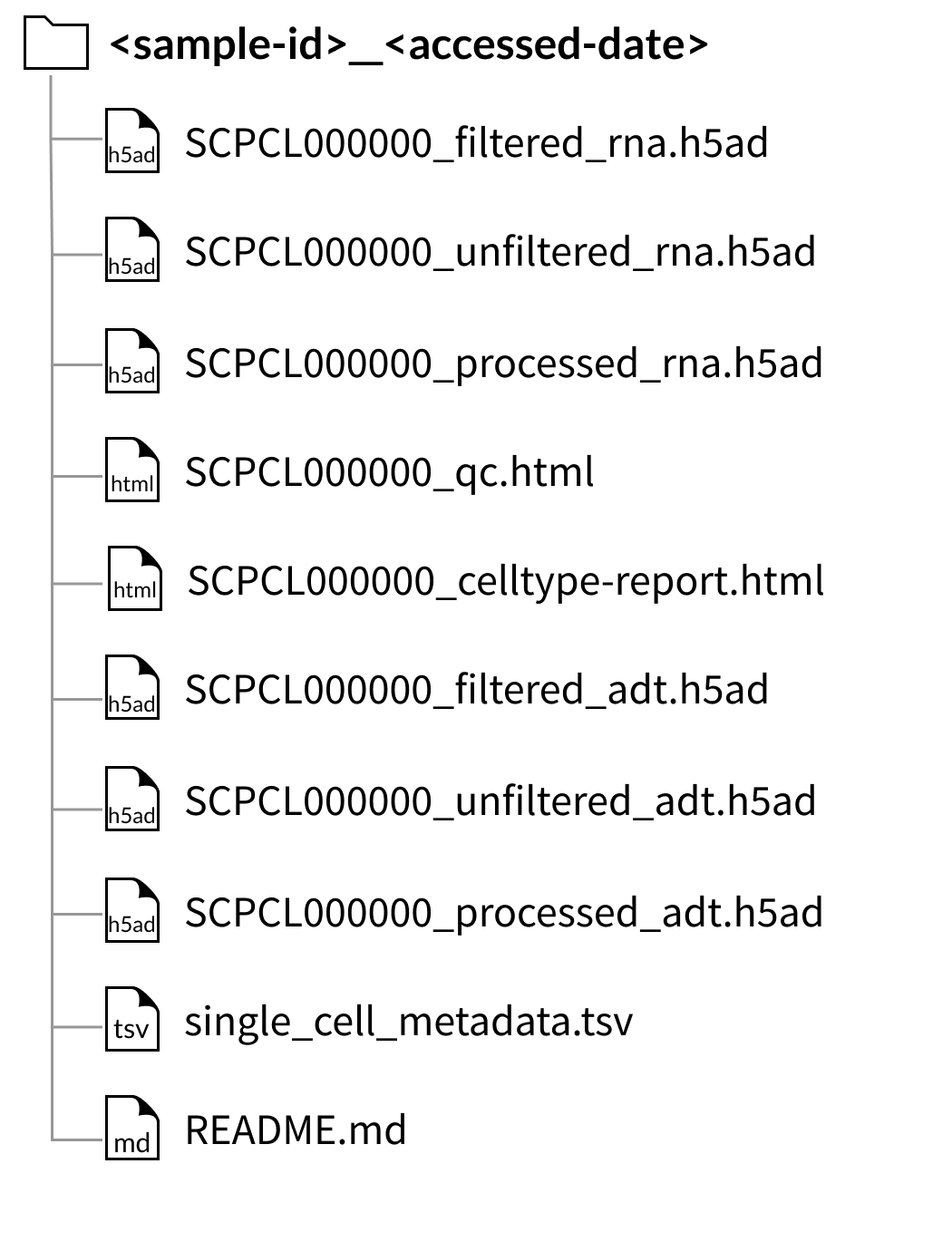
AnnData downloads
Download folder structure for project downloads:
Download folder structure for individual sample downloads:
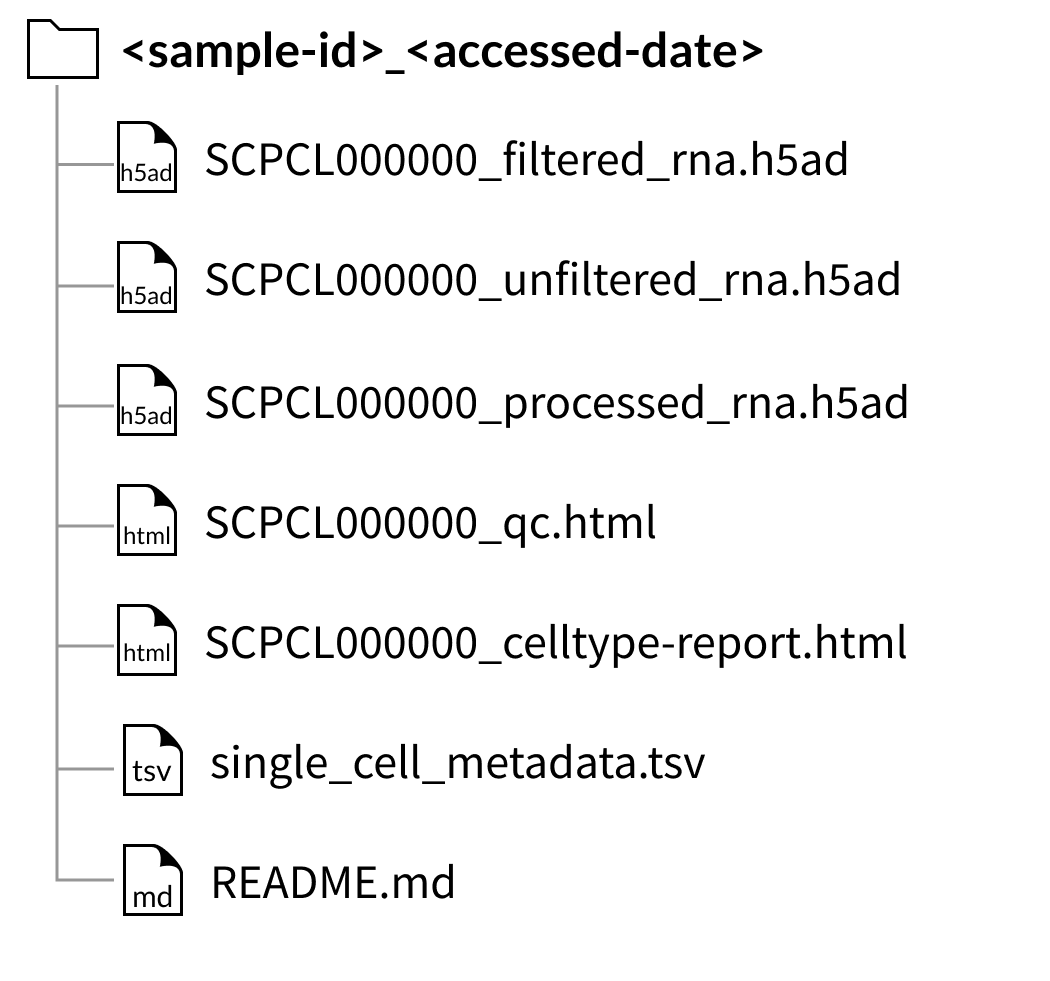
Download folder structure for individual sample downloads with CITE-seq (ADT) data:
If downloading a sample that contains a CITE-seq library as an AnnData object (hdf5 file), the quantified CITE-seq expression data is included as a separate file with the suffix _adt.hdf5.
Gene expression data
Single-cell or single-nuclei gene expression data is provided as either SingleCellExperiment objects (.rds files) or AnnData objects (.hdf5 files).
Three files will be provided for each library included in the download - an unfiltered counts file, a filtered counts file, and a processed counts file.
The unfiltered counts file, SCPCL000000_unfiltered.rds or SCPCL000000_unfiltered_rna.hdf5, contains the counts matrix, where the rows correspond to genes or features and the columns correspond to cell barcodes.
Here, all potential cell barcodes that are identified after running alevin-fry are included in the counts matrix.
The object also includes summary statistics for each cell barcode and gene, as well as metadata about that particular library, such as the reference index and software versions used for mapping and quantification.
The filtered counts file, SCPCL000000_filtered.rds or SCPCL000000_filtered_rna.hdf5 contains a counts matrix with the same structure as above.
The cells in this file are those that remain after filtering using emptyDrops.
As a result, this file only contains cell barcodes that are likely to correspond to true cells.
The processed counts file, SCPCL000000_processed.rds or SCPCL000000_processed_rna.hdf5, contains both the raw and normalized counts matrices.
The filtered counts file is further filtered to remove low quality cells, such as those with a low number of genes detected or high mitochondrial content.
This file contains the raw and normalized counts data for cell barcodes that have passed both levels of filtering.
In addition to the counts matrices, the SingleCellExperiment or AnnData object stored in the file includes the results of dimensionality reduction using both principal component analysis (PCA) and UMAP.
See Single-cell gene expression file contents for more information about the contents of the SingleCellExperiment and AnnData objects and the included statistics and metadata.
See also Using the provided RDS files in R and Using the provided HDF5 files in Python.
QC report
The included QC report, SCPCL000000_qc.html, serves as a general overview of each library, including processing information, summary statistics and general visualizations of cell metrics.
Cell type report
The cell type report, SCPCL000000_celltype-report.html, includes an overview of cell type annotations present in the processed objects.
This report contains details on methodologies used for cell type annotation, information about reference sources, comparisons among cell type annotation methods, and diagnostic plots.
For more information on how cell types were annotated, see the section on Cell type annotation.
If the downloaded library was from a cell line sample, no cell type annotation will have been performed. Therefore, there will be no cell type report in the download for these libraries.
Metadata
The single_cell_metadata.tsv file is a tab-separated table with one row per library and the following columns.
column_id |
contents |
|---|---|
|
Sample ID in the form |
|
Library ID in the form |
|
|
|
10x kit used to process library |
|
Number of cells after filtering with |
|
Original sample identifier from submitter |
|
Unique id corresponding to the donor from which the sample was obtained |
|
Submitter name/id |
|
Age at time sample was obtained |
|
Sex of patient that the sample was obtained from |
|
Tumor type |
|
Subcategory of diagnosis or mutation status (if applicable) |
|
Where in the body the tumor sample was located |
|
At what stage of disease the sample was obtained, either diagnosis or recurrence |
|
The organism the sample was obtained from (e.g., |
|
|
|
|
|
NCBI taxonomy term for organism, e.g. |
|
For Homo sapiens samples, a |
|
|
|
|
Additional metadata may also be included, specific to the disease type and experimental design of the project.
Examples of this include treatment or outcome.
Metadata pertaining to processing will also be available in this table and inside of the SingleCellExperiment object.
See the SingleCellExperiment experiment metadata section for more information on metadata columns that can be found in the SingleCellExperiment object.
See the AnnData experiment metadata section for more information on metadata columns that can be found in the AnnData object.
For projects with bulk RNA-seq data, the bulk_metadata.tsv file will be included for project downloads.
This file will contain fields equivalent to those found in the single_cell_metadata.tsv related to processing the sample, but will not contain patient or disease specific metadata (e.g. age, sex, diagnosis, subdiagnosis, tissue_location, or disease_timing).
Multiplexed sample libraries
For libraries where multiple biological samples were combined via cellhashing or similar technology (see the FAQ section about multiplexed samples), the organization of the downloaded files and metadata is slightly different.
Note that multiplexed sample libraries are only available as SingleCellExperiment objects, and are not currently available as AnnData objects.
For project downloads, the counts and QC files will be organized by the set of samples that comprise each library, rather than in individual sample folders.
These sample set folders are named with an underscore-separated list of the sample ids for the libraries within, e.g., SCPCS999990_SCPCS999991_SCPCS999992.
Bulk RNA-seq data, if present, will follow the same format as bulk RNA-seq for single-sample libraries.
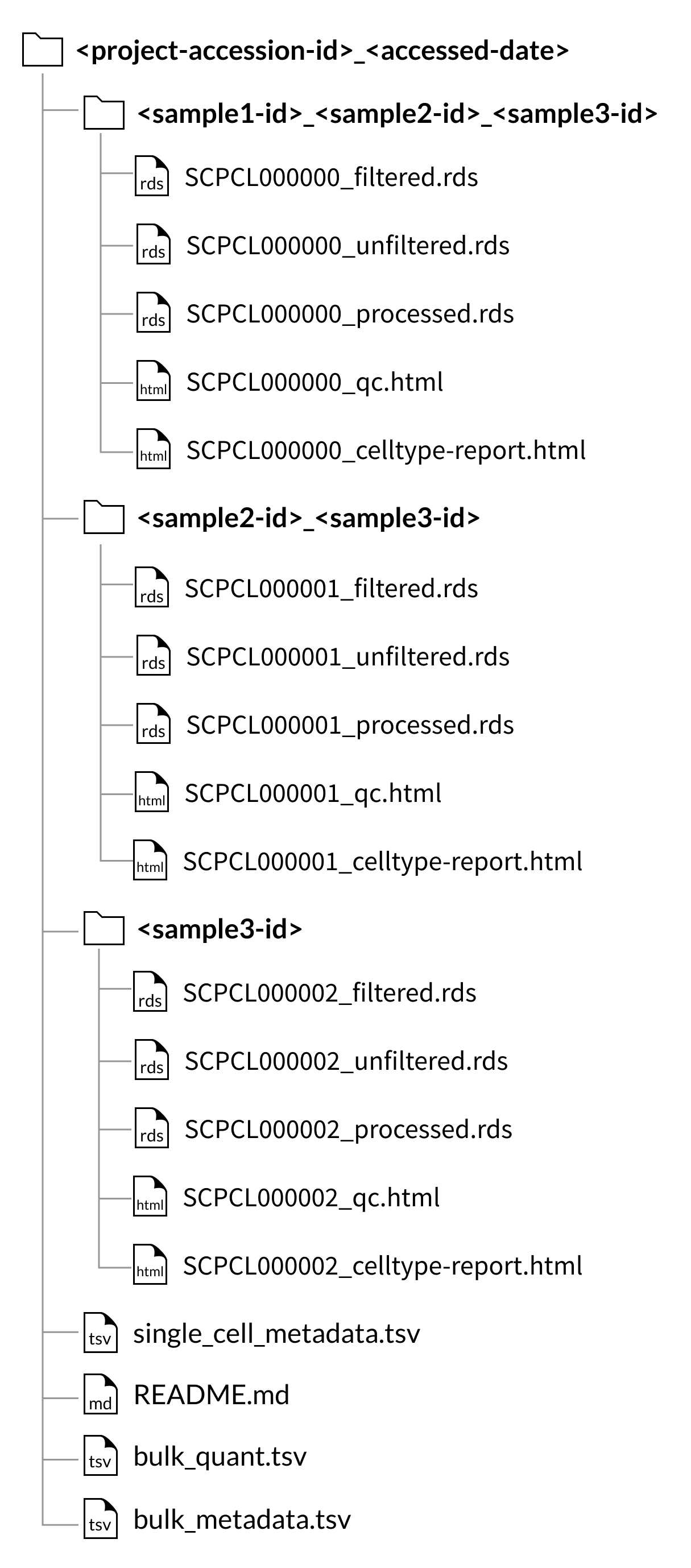
Because we do not perform demultiplexing to separate cells from multiplexed libraries into sample-specific count matrices, sample downloads from a project with multiplexed data will include all libraries that contain the sample of interest, but these libraries will still contain cells from other samples.
For more on the specific contents of multiplexed library SingleCellExperiment objects, see the Additional SingleCellExperiment components for multiplexed libraries section.
The metadata file for multiplexed libraries (single_cell_metadata.tsv) will have the same format as for individual samples, but each row will represent a particular sample/library pair, meaning that there may be multiple rows for each scpca_library_id, one for each scpca_sample_id within that library.
Merged object downloads
When downloading a full ScPCA project, you can choose to download data from all samples as individual files, or you can download a single file containing all samples merged into a single object.
Merged object downloads contain all single-cell or single-nuclei gene expression data for a given ScPCA project within a single object, provided as either a SingleCellExperiment object (.rds file) or an AnnData object (.hdf5 file).
The object file, SCPCP000000_merged.rds or SCPCP000000_merged_rna.hdf5, contains both a raw and normalized counts matrix, each with combined counts for all samples in an ScPCA project.
In addition to the counts matrices, the SingleCellExperiment or AnnData object stored in the file includes the results of library-weighted dimensionality reduction using both principal component analysis (PCA) and UMAP.
See the section on merged object processing for more information about how merged objects were created.
If downloading a project that contains at least one CITE-seq library, the quantified CITE-seq expression data will also be merged.
In SingleCellExperiment objects (rds files), the CITE-seq expression data is provided as an alternative experiment in the same object as the gene expression data.
However, for AnnData objects, (hdf5 files), the quantified CITE-seq expression is instead provided as a separate file called SCPCP000000_merged_adt.hdf5.
Every download also includes a single single_cell_metadata.tsv file containing metadata for all libraries included in the merged object.
For a full description of this file’s contents, refer to the metadata section above.
If downloading a project containing bulk RNA-seq data, two tab-separated value files, bulk_quant.tsv and bulk_metadata.tsv, will be included in the merged object download.
The bulk_quant.tsv file contains a gene by sample matrix (each row a gene, each column a sample) containing raw gene expression counts quantified by Salmon.
The bulk_metadata.tsv file contains associated metadata for all samples with bulk RNA-seq data.
This file will contain fields equivalent to those found in the single_cell_metadata.tsv related to processing the sample, but will not contain patient or disease specific metadata (e.g. age, sex, diagnosis, subdiagnosis, tissue_location, or disease_timing).
Every download includes a summary report, SCPCL000000_merged-summary-report.html, which provides a brief summary of the samples and libraries included in the merged object.
This includes a summary of the types of libraries (e.g., single-cell, single-nuclei, with CITE-seq) and sample diagnoses included in the object, as well as UMAP visualizations highlighting each library.
Every download also includes the individual QC report and, if applicable, cell type annotation reports for each library included in the merged object.
Download folder structure for SingleCellExperiment merged downloads:
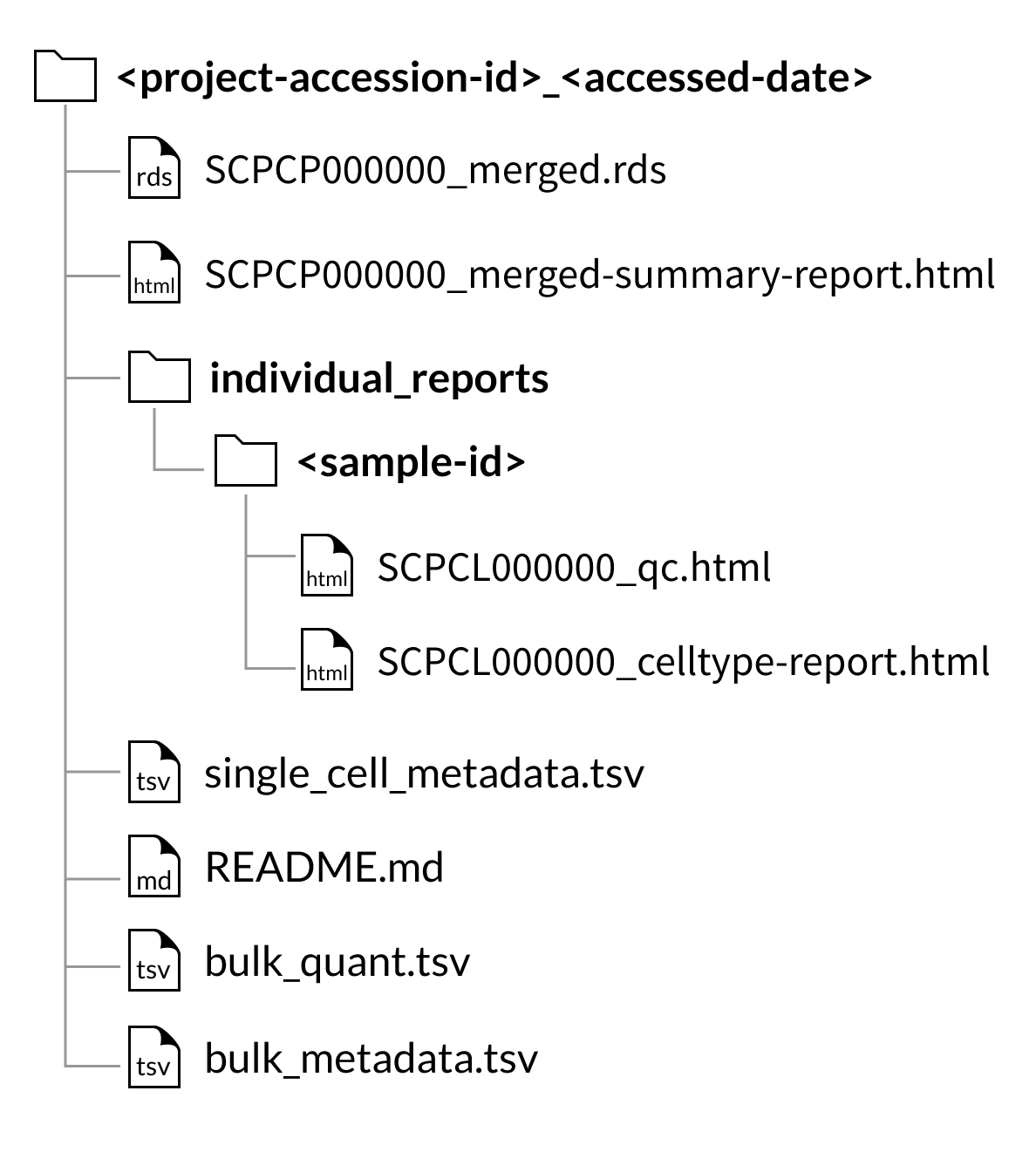
Download folder structure for AnnData merged downloads:
Download folder structure for AnnData merged downloads with CITE-seq (ADT) data:
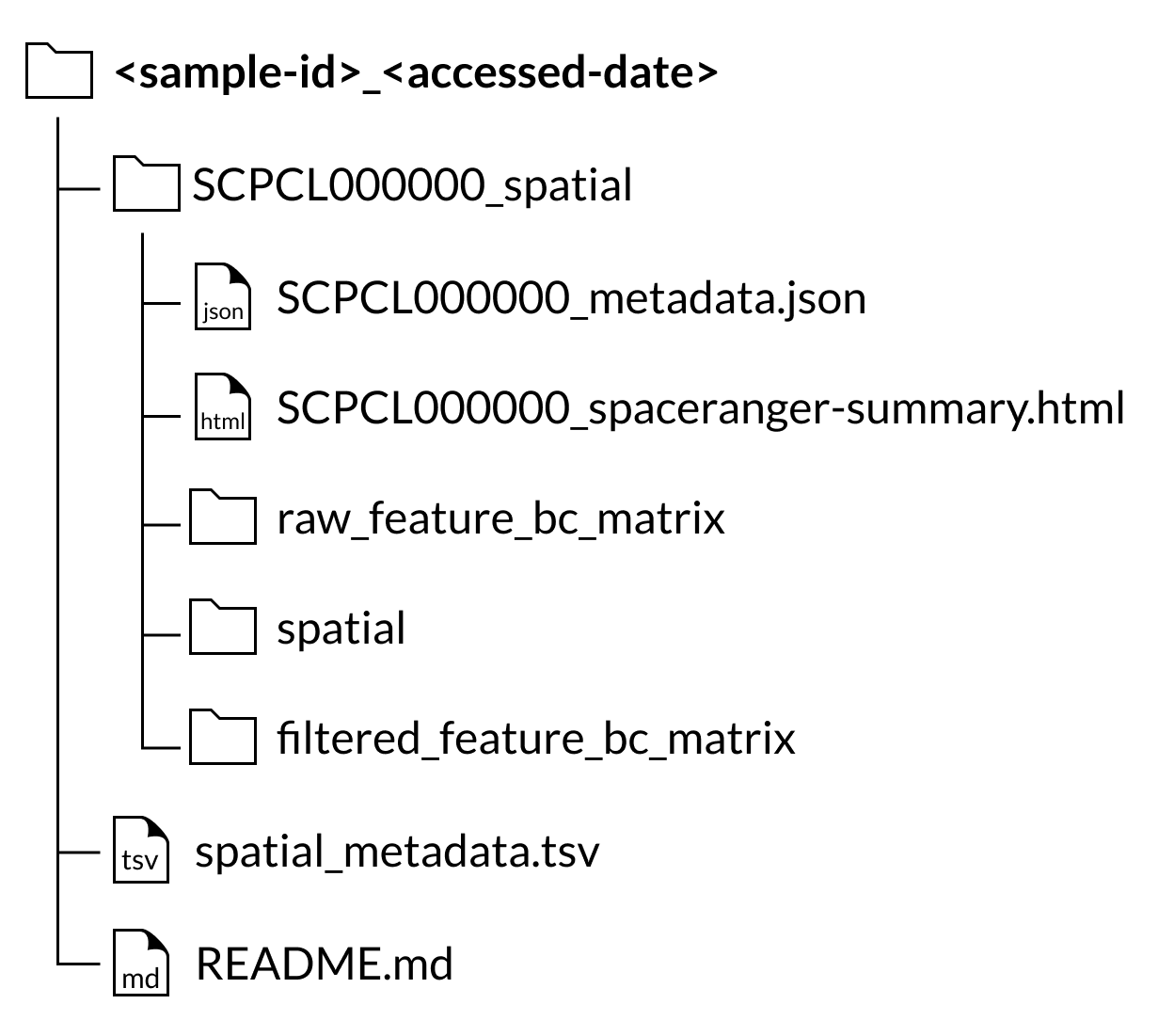
Spatial transcriptomics libraries
If a sample includes a library processed using spatial transcriptomics, the spatial transcriptomics output files will be available as a separate download from the single-cell/single-nuclei gene expression data.
For all spatial transcriptomics libraries, a SCPCL000000_spatial folder will be nested inside the corresponding sample folder in the download.
Inside that folder will be the following folders and files:
A
raw_feature_bc_matrixfolder containing the unfiltered counts filesA
filtered_feature_bc_matrixfolder containing the filtered counts filesA
spatialfolder containing images and position informationA
SCPCL000000_spaceranger-summary.htmlfile containing the summary html report provided by Space RangerA
SCPCL000000_metadata.jsonfile containing library processing information.
A full description of all files included in the download for spatial transcriptomics libraries can also be found in the spaceranger count documentation.
Every download also includes a single spatial_metadata.tsv file containing metadata for all libraries included in the download.